Hi everyone,
Hi everyone,
The eagerly awaited OpenPCR kit is now shipping! UPS picked up the first batch of kits and OpenPCRs are on their way to users in 5 continents and 13 countries around the world. For $512, every OpenPCR kit includes all the parts, tools, and beautiful printed instructions – you ONLY need a set of screwdrivers.
A PCR machine is basically a copy machine for DNA. It is essential for most work with DNA, things like exposing fraud at a sushi restaurant, diagnosing diseases including HIV and H1N1, or exploring your own genome. The guy who discovered the PCR process earned a Nobel Prize in 1993, and OpenPCR is now the first open source PCR machine.
The price of a traditional PCR machine is around $3,000. So, do people in garages have great PCR machines? Not really. Howabout high school or middle school teachers? Nope. Howabout smaller medical testing labs or labs in India or China? Nope. Even some big bio labs try their luck on eBay. We set out to change that.
Josh and I prototyped OpenPCR over about 4 months — it was a lot of fun. Last May we unveiled the first OpenPCR prototype to all a bunch of crazy people on Kickstarter, 158 people gave us a total of $12,121. With that we designed and manufactured a repeatable, works-all-the-time device — it took a lot of hard work. Now we’re done and ready to share!
OpenPCR Firsts:
1. First commercially available PCR machine for $512
We get a lot of people who come up to us and say “jumping jillikers, batman! we paid $10,000 for ours and it’s this big (make refrigerator-sized hand motion)!”. While modern PCR machines aren’t fridge sized anymore, we’re proud to say that OpenPCR is the most affordable and most compact PCR machine out there.
2. First Arduino USB storage device:
This is a big deal for you Arduino hackers out there. A normal Arduino can only talk back and forth over a serial port. This is a pain to set up, and we wanted OpenPCR to just plug-in and go. How does it work? When OpenPCR is plugged in, the Arduino mounts itself as a USB drive called “OpenPCR”. The computer passes love notes to OpenPCR by writing to that file, and Arduino sends love notes back by writing to another file. The implementation was tough, and there are size restrictions due to the size of the chips used by Arduino, but it’s pretty simple to make use of. We also built a cross-platform app for your Mac or PC in Adobe Air so that the we could have a simple computer control interface. Simply plug in your OpenPCR to your computer with USB. No setup besides downloading the OpenPCR app! (Josh and Xia totally pulled off a miracle on this!) If you’ve got questions on this specifically, be sure to post below!
No cutting corners
The clear vision of OpenPCR that made it great was driven by 2 things. First off, Josh is an incredible engineer and we both enjoyed learning a lot of new things over the past year — everything from how to make circuit boards, machine metal parts, laser cutting, Arduino hacking, USB hacking. I’d say 90% of the success of OpenPCR was lots of hard work. Hard work is great but there are lots of projects where hard work is put in but never “pays off”. How did we stay on course? I think the prototype + showing it off on Kickstarter/Maker Faire had a lot to do with it. We of course had lots of exciting ideas about new functionality and extra things over the past year. The beauty of having built our prototype was we knew if we could just get to that point we would have a hit.
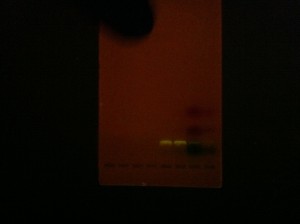
 For example, we designed OpenPCR to be assembled by hand. The printed Build Instructions are a big part of OpenPCR and we did a lot of work to get them right. As we finalized the OpenPCR design a few steps stood out as “hard”. We switched from thermal paste to thermal pads (not messy, no need for gloves), assembled circuit boards (no need for a pro soldering setup), and pre-epoxied the thermistor. The OpenPCR kit is easy to build because of those decisions. We’ve still got to publish the gel pictures showing how great OpenPCR works, but that’s been well tested ourselves. If you’ve got an OpenPCR kit coming your way and would like to post pictures of a gel run afterwards, we would love to see your results too!
For example, we designed OpenPCR to be assembled by hand. The printed Build Instructions are a big part of OpenPCR and we did a lot of work to get them right. As we finalized the OpenPCR design a few steps stood out as “hard”. We switched from thermal paste to thermal pads (not messy, no need for gloves), assembled circuit boards (no need for a pro soldering setup), and pre-epoxied the thermistor. The OpenPCR kit is easy to build because of those decisions. We’ve still got to publish the gel pictures showing how great OpenPCR works, but that’s been well tested ourselves. If you’ve got an OpenPCR kit coming your way and would like to post pictures of a gel run afterwards, we would love to see your results too!
The intent of the prototype was simple – we wanted a PCR machine for people like us. That meant a 16 well PCR machine controlled by computer, with a built in screen, good for the lab bench or a workshop/garage. And that’s exactly what OpenPCR is.
Where did the time go?
After Kickstarter started in May, we worked for going on 14 months now. Between Josh and I, I estimate we put about 3,000 hours into OpenPCR, not counting the time leading up to the prototype. We’ve got 57 posts and 600+ comments on the OpenPCR blog, covering a lot of aspects of OpenPCR development. In the past few months we’ve kept our heads down getting everything out the door and we’ve got some stories to share. Short answer is, there’s a lot of blogging to catch up on.
Special thanks to Xia Hong, Eri Gentry, and Will Reinhardt who volunteered lots of their time to help OpenPCR.

Just the beginning
OpenPCR is designed for labs, classrooms, and garages. Tell your science-y friends about OpenPCR, “Like” us on Facebook, or write us and tell us that you stopped by! You can also get your own OpenPCR kit!
Do you want to see us develop more breakthrough biotechnology? Along this journey we uncovered a lot of opportunities for PCR and other biological devices. We’re a new company and would love to meet other passionate people. Our hurdles right now are manufacturing (mechanical engineers!), distribution (sales + marketers!), and new hardware (hackers!)/software (hackers!)/bioware (biologists!) + industrial design. If you’re in the Bay Area and want to get in on making all this crazy DNA stuff useful to regular people, send us an email: contact@openpcr.org.
For more information, we’ve gotten a lot of media attention over the past year including NYTimes, GQ France, Biotechniques, and USA Today.
Ordered a kit and wondering where it is? We have shipped a first batch of kits and emailed out tracking numbers to the recipients. If your kit hasn’t shipped yet, we’re working on shipping a second batch and will keep you updated.
[button color="blue" size="large" href="https://openpcr.org/buy" ]Buy Now[/button]